
The structure and molecular components of biological membranes allow
Chapter 14, pages 160-161, 447-462.
To explain the use of centrifugation to isolate and characterize cell fractions.
To explain the use of autoradiography to follow labelled cell components through space and time.
Think about the postal address analogy . One should be able to explain how a protein (e.g. procollagen) gets from the point of synthesis on rough ER to its free (or biologically active) state on the outside of the cell.
Alternatively, one should be able to explain how a protein gets from its site of synthesis to the inside of mitochondrion, or chloroplast, lysosome or the nucleus.
At each step there is information built into both the protein and the target.
As molecules move from the RER to the outside of the cell, they are tested at every step of the way. Only molecules with the correct signals are allowed to proceed.
Look carefully at questions above. Think about possible exam questions. Remember that process questions are a 'stock in trade' of those who set final exam questions in courses such as this one.
Two sections to this lecture:
1. How to study organelles.
2. Explanation of three major means for protein to be imported into organelles.
 |
Look at Figure 14-2. The cell is filled with a large number of membrane limited compartments (membrane limited spaces as well as the membranes themselves). |
How to study these cellular compartments?
1. Visual techniques
Microscopy of fixed material - shows what is there, but presents a static view. May be light or election microscopic analysis.
Technical uses of microscopy - for example, phase contrast, immuno-labeling techniques, fluorescence microscopy etc.
Staining - specific components may be stained. e.g. Janus green stains mitochondria in live cells. Phloroglycinol (sp?) stains lignin in plant cell walls. More recently a wide array of immuno-labeling techniques have come into use that make it possible to follow specific proteins. These are very important procedures because they make it possible to follow individual proteins.
2. Biochemistry.
Single cell chemistry - done initially on yeasts, bacteria and protists.
Blender biochemistry (Grind and Find) - cells are ground up in a blender (technically called a homogenizer, but the same thing as a kitchen blender). This made the discovery of a lot of enzyme chemistry (for example glycolysis) possible. One of the problems is that the preparations are labile because cell process are dynamic and will not work long when the system is disrupted.
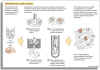 Cell homogenization (Ya gotta grind before ya can find!)
Cell homogenization (Ya gotta grind before ya can find!)
Biochemical tests on microscopic samples (cytochemistry). The principle here is to do a chemical test that can be interpreted by microscopic examination. This is useful for telling where in the cell a reaction is occurring. Example - use of tetrazolium salts to detect reduction/ oxidization reactions, for example in mitochondria.
How do proteins get from the rough endoplasmic reticulum to the outside of the cell? How to study this.
How can this be used to track a protein in transit?
Continuous labelling experiment. Feed a radio-labelled precursor (e.g. 35S-Methionine), then lyse and homogenize cells after various amounts of time have passed (30 sec, 1 min, 2 min, 5 min, 10 min.). Sample, homogenize, centrifuge, assay. Look for label in the various fractions. Look for the protein of interest, e.g. (procollagen).
Pulse-Chase experiment. An important variant of this procedure is the pulse - chase experiment. The objective is to produce a population of highly labelled proteins that were all synthesized at the same time. These may then be followed through subsequent processing steps in the cell. This experiment produces information about where the molecules go and when they get there. Imagine that you wanted to find out what happened to high school graduates in their subsequent lives. You might select a representative sample of graduates, say the Lord Byng class of 1999 (imagine them all painted red), and follow them over time. This experiment does the same thing with molecules. In this procedure cells are gives a short exposure to a high concentration of radioactively labelled precursor (e.g. 3H-leucine).This is the pulse part of the experiment. This is then followed by removal of the labelled medium, if possible, and the administration of a great excess of non-labelled leucine. This addition of non-labelled amino acid greatly reduces the specific activity of the leucine in the cell. This is the chase part of the experiment. The key idea is that all of the labelled molecules were made during the few minutes that the high activity label was present in the system. Their radioactive label allows them be be picked out against a background of non-labelled molecules of a wide variety of ages, both older and younger.
Cell fractionation can be used to follow labelled molecules in a pulse chase experiment. That is, you sample the population at different times after the start of the chase (e.g. 3 min, 5 min, 10, 15, 20, 30, 60 min), then lyse the cells and separate the bits of ER, Golgi, Secretory granules, extracellular material etc by differential centrifugation and other biochemical procedures. You then them determine how much time elapses before label appears in secretory granules, for example. Click on the figure below for an example.
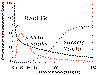 Time
course of movement of a labelled protein through compartments in a secretory
cell. Click on thumbnail to enlarge. This type of information can be obtained
by cell fractionation or by autoradiography. Time
course of movement of a labelled protein through compartments in a secretory
cell. Click on thumbnail to enlarge. This type of information can be obtained
by cell fractionation or by autoradiography. |
Autoradiography is an alternative approach to the study of labelled proteins through the secretory process. |
| This diagram shows the time course of a labelled protein as it is processes for the secretory process. In this experiment acinar cells of the pancreas were given a tritium-(3H) labelled amino acid which was incorporated into proteins during the first minutes of the experiment. The labelled amino acid was then replaced by a large excess of non-labelled amino acid (chased), the the labelled material already incorporated into proteins was followed through the secretory process. Notice that it quickly leaves the endoplasmic reticulum (0 to 5 min), passes through the Golgi vesicles (7-20 min) and accumulates in secretory vesicles (50-120 min). | |