Flow cytometry (FCM) is – in principle – a relatively simple technique that allows one to measure properties of cells or organelles as they are intercepted (individually) by a high-intensity light source. In plant biology, it is often used to analyze extracts of intact nuclei, with the aim of determining DNA content and (indirectly) ploidy levels (this is the application I will focus on here). A good reference (including troubleshooting tips) can be found here.
Illumina Sequencing Adapters (Dan E.)
I’ve been writing posts about the various steps involved in making WGS sequencing libraries for the Biodiversity Centre’s Illumina HiSeq machine and I was tempted to try to explain what the adapters are for. Then I had the bright idea of seeing if somebody else had already done it. Somebody has. Continue reading
Illumina Sequencing Adapters and Barcodes (Dan E.)
As of March 2012 we are using the Bioo Scientific NEXTflex barcoded adapters for WGS sequencing libraries made by ourselves, (well me so far). The set we are currently using comprises 48 barcodes, so we can multiplex up to a 48-plex in one lane on the Illumina HiSeq sequencer.
Below are the sequences of the Illumina adapters and the 48 barcodes we are currently using. Continue reading
For your free time (Kathryn)
Phylo: crowd-sourced tetris-like game for genomic sequence alignment. Pretty sweet. Contribute to science! You know, even more than you already do…
Phylo Tutorial from Phylo Game on Vimeo.
Ampure XP beads – DNA clean up and size selection (Dan E.)
Ampure XP magnetic beads have become popular for cleaning DNA from reactions and for size selecting DNA. WGS sequencing library preparation protocols, and the GBS protocol, involve one or more bead clean up steps. If you make next gen sequencing libraries you will use Ampure XP beads. Following is some basic information about them and my thoughts about using them, mainly for library preparations . . . Continue reading
Using RSEM to estimate gene and isoform expression (Sam)
RSEM is a relatively new bioinformatics tool that has been developed in conjunction with Trinity for the analysis of RNAseq data. RSEM can be used to estimate expression levels for both genes and different isoforms of genes, and is quite quick and easy to use, with an excellent google group for help (“RSEM users”). All it requires is an RNAseq dataset (either fasta or fq format) and a reference transcriptome that it can be aligned to.
Watch out for this (Kathryn)
Horticulture Greenhouse Space (Kate)
Our lab is currently paying for one bench in the Horticulture greenhouse for general purposes (Zone 4, Bench #1). This bench is intended for small, short-term projects only. If you need more than ~1/3 of a bench, please make your own arrangements.
About the space: . . . Continue reading
GNU Parallel (Chris)
GNU Parallel makes it easy to take advantage of multiple processors at once. I really enjoy using it to parallelize Bash scripts. For example, here’s a pipeline that will generate a reference-based transcriptome from Illumina-sequenced ESTs: . . . Continue reading
Tissue area measurements with ImageJ (Allan)
See also this post by Kathryn (Dan E.).
Goal. Convert area to pixels. Measure each pixel for precise area measurement of odd shapes (leaves, seeds etc.) without bias. I’ve included a practice image of an Arabidopsis stem. Continue reading
Loading Dye for Agarose Gels (Dan E.)
If you are new to the lab bench, and perhaps even if you’re not, you may not know how to make loading dye for agarose gels. Its the sort of thing that tends to get made rarely and then persist, so people often inherit a supply and have no idea how to make more when they run out. For this sort of thing the best reference is the relevant appendix at the back of the Sambrook et al. Molecular Cloning text books (near Megan’s desk) and, of course, there is Google. Or, you can just do what I do. My recipe is incredibly simple and would be close to the cheapest option too. Continue reading
Gel doc Dropbox (Allan)
Hi all,
I’m not sure how most of you were sharing your gels but I set up a dropbox folder within the Rieseberg gels folder. Sending e-mails was tiresome. If you’re interested in synchronizing your gels with your computer drag your folder(s) to the dropbox folder. The dropbox account credentials (online and client) are as follows:
user=allandebono@allandebono.org
password=1d7hz@!
… Go through the process as usual to set up a shared folder.
Gel Slice – size selection or band isolation (Dan E.)
Cutting a slice from an agarose gel in order to size select DNA or to isolate a PCR product etc. is standard practice in lab genetics. This post is about how I do it in our lab, (as of Feb 2012).
There are three features of my current method that may be unfamiliar to you even if you’ve done this sort of thing before:
- The Dark Reader transilluminator.
- SYBR-Gold.
- Sigma X-Tracta gel punches.
Continue reading
Protocol: Whole Genome Amplification using Qiagen REPLI-g ultra fast kit (Moira)
Here is the WGA Qiagen REPLIg ultra fast PROTOCOL.
I added also general information (mostly from the Qiagen site) about the REPLI-g method of amplification and about other similar Qiagen kits available.
Continue reading
Turning your SNP table into a STRUCTURE input file (Brook)
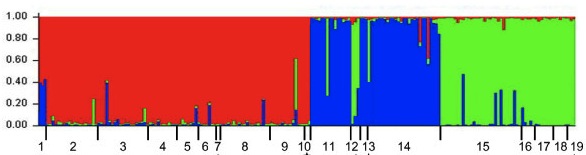 I suspect there are probably several homemade versions of this kind of script kicking around, but here is a perl script I’ve written for turning your SNP table into a STRUCTURE input file. To use it, you should change the .txt to a .pl after downloading the script. More on STRUCTURE input files (and so much more!) is in the documentation here.
I suspect there are probably several homemade versions of this kind of script kicking around, but here is a perl script I’ve written for turning your SNP table into a STRUCTURE input file. To use it, you should change the .txt to a .pl after downloading the script. More on STRUCTURE input files (and so much more!) is in the documentation here.
How to log in to RLR (Dan E.)
Hello,
If you are a Riesebergler and a member of RLR you will have a user name and password that you can enter into the log in boxes at the top right of the Home page or at this page to log in to RLR.
If you have not logged in you will see only those posts published publicly. After you log in you will also be able to see all posts that have been published privately (post titles start with “Private: “).
Dan.
Projector Tricks (Kate)
Below are settings that appear to get my computer (a MacBook) to communicate properly with the lab projector. I don’t promise that these settings will work for any other computer.
System preferences -> Displays
This will bring up two windows. One that corresponds to your computer screen and one that corresponds to the projector (see the top of the windows to determine which is which). Note that you can use the gather windows button to bring the projector window on to your computer screen.
Under the arrangement tab on the computer screen window uncheck mirror displays.
Under the display tab on the projector window choose 1024 x 768.
Note that choosing a widescreen resolution for the projector (e.g. 1600 x 900) results in a cut-off picture even though the projector claims to be widescreen.
I hope this is helpful!
Kate
If BWA wants *.nt.ann file… (Rose)
Recently BWA (an alignment program) suddenly started giving a strange error message, indicating that a reference file ending in *.nt.ann was missing. This file type was unfamiliar to me, with good reason: it’s a colourspace reference file, which shouldn’t be generated when we index the fasta-based references we’re using (at least, I don’t know of anyone in our lab using SOLID data as a reference). DO NOT rebuild the reference with the -c (colourspace) flag, as you might see suggested on the web, because we don’t know what effect that might have on our alignments. DO rebuild it with the usual settings.
Unix cheat sheet (Dan E.)
Hi guys,
Following up on Nolan’s bioinfo workshop last week, here is a copy of
Alistair’s cheat sheet from the workshop he gave last year. I have
found it quite useful.
cheers,
seb
Zymo Research Genomic DNA Clean & Concentrator Columns (Dan E. and Kate)
We have some columns in the lab that are for cleaning and concentrating genomic DNA. They are made by an outfit called Zymo Research and should be easy to find, look for a bright yellow box.
This post describes a test I ran of these columns. Continue reading