The “protocol” that we have been using is available here as a Google doc. I use quotes there because, as you will see, there is no single protocol used by the lab to date. Without analyzed sequence data yet to compare the described protocols, the best advice I can give to you is to pick a sensible pipeline for your needs, taking into consideration the time/effort/desired output that works for you, and apply that pipeline consistently to all samples for the same project. All sets of methods described have resulted in libraries that pass QC and generate a sufficient reads during sequencing. Good luck!
Author Archives: Brook
Advice for the poor resident of Vancouver
Hello everyone! The Botany Graduate Student Association recently put together a couple of factsheets dealing with financial matters as a graduate student at UBC. Some of the advice is specific to students, but some of it could be useful to any new resident of Vancouver struggling to deal with the cost of living here. The factsheets are available via the link above.
PstI-MspI GBS: an update
How my PhD was saved by DTT, or: how to get moderate quantities of clean, unsheared DNA from tricky tissues
I have been trying for months to find a high-throughput solution to DNA extraction from lyophilized (freeze-dried) leaf discs of Helianthus argophyllus, a species known for its intransigent polyphenolics and low DNA yields. And I appear to have found one, thanks to Horne et al. 2004 (here) and Dow Chemical!
“Lab” bike
Summer is almost upon us, and many of you might be considering taking field trips to help out down at Totem or the Farm. So I’d thought I’d let you know about my crappy field bike, available for use to get yourself around campus!
lab contact information and birthday list
Here is the most up-to-date contact information for the Rieseberg lab, including birthdays, email addresses, and phone numbers. If you want, you can download a word document of this here: Lab Contact Info and Birthday List. If your information changes, either let me know or edit this post and the downloadable document appropriately. Thanks!
(contact: John L)
Guide to Next-Generation Sequencers (Brook)
Occasionally I find myself reading a news item or a paper that mentions a particular sequencing platform and scratching my head to remember of what exactly that particular platform is capable. If you ever find yourself in that same boat, the Molecular Ecologist has a very handy and often-updated guide here.
Turning your SNP table into a STRUCTURE input file (Brook)
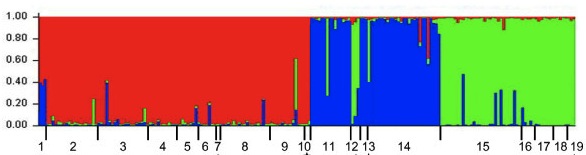 I suspect there are probably several homemade versions of this kind of script kicking around, but here is a perl script I’ve written for turning your SNP table into a STRUCTURE input file. To use it, you should change the .txt to a .pl after downloading the script. More on STRUCTURE input files (and so much more!) is in the documentation here.
I suspect there are probably several homemade versions of this kind of script kicking around, but here is a perl script I’ve written for turning your SNP table into a STRUCTURE input file. To use it, you should change the .txt to a .pl after downloading the script. More on STRUCTURE input files (and so much more!) is in the documentation here.
Macro Phenotyping (Brook)
 This post is meant to be a mostly comprehensive list and explanation of the non-molecular phenotypes that our lab measures on our various plant populations. At the moment, it is definitely not there yet (for one thing, it has a clear sunflower bias), so please contribute! Continue reading
This post is meant to be a mostly comprehensive list and explanation of the non-molecular phenotypes that our lab measures on our various plant populations. At the moment, it is definitely not there yet (for one thing, it has a clear sunflower bias), so please contribute! Continue reading