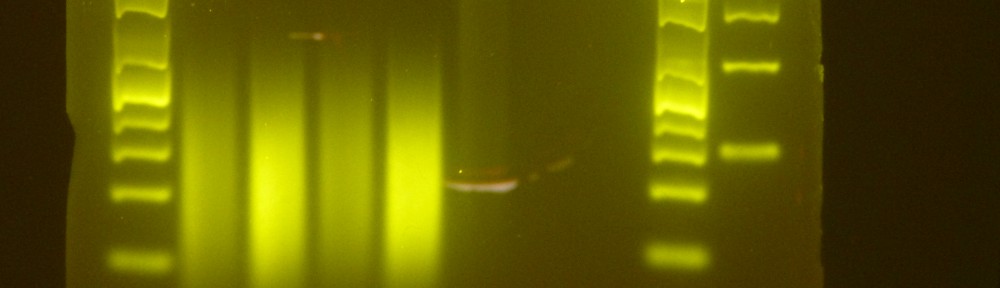
The “protocol” that we have been using is available here as a Google doc. I use quotes there because, as you will see, there is no single protocol used by the lab to date. Without analyzed sequence data yet to compare the described protocols, the best advice I can give to you is to pick a sensible pipeline for your needs, taking into consideration the time/effort/desired output that works for you, and apply that pipeline consistently to all samples for the same project. All sets of methods described have resulted in libraries that pass QC and generate a sufficient reads during sequencing. Good luck!
Rieseberg Lab Resources
RLR: Technical resources for Rieseberglers