Congratulations to PhD Candidate Rashika Ranasinghe and coauthor Dr. Sampath Seneviratne on today’s publication of this paper:
Ranasinghe, R.W., S.S. Seneviratne, and D. Irwin. 2024. Cryptic hybridization dynamics in a three-way hybrid zone of Dinopium Flamebacks on a Tropical Island. Ecology and Evolution 14: e70716, https://doi.org/10.1002/ece3.70716
The Abstract:
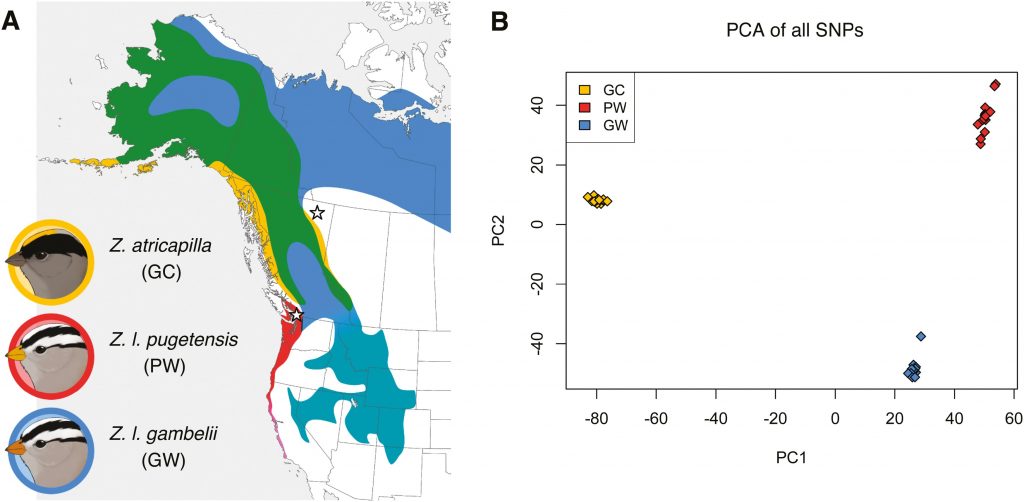
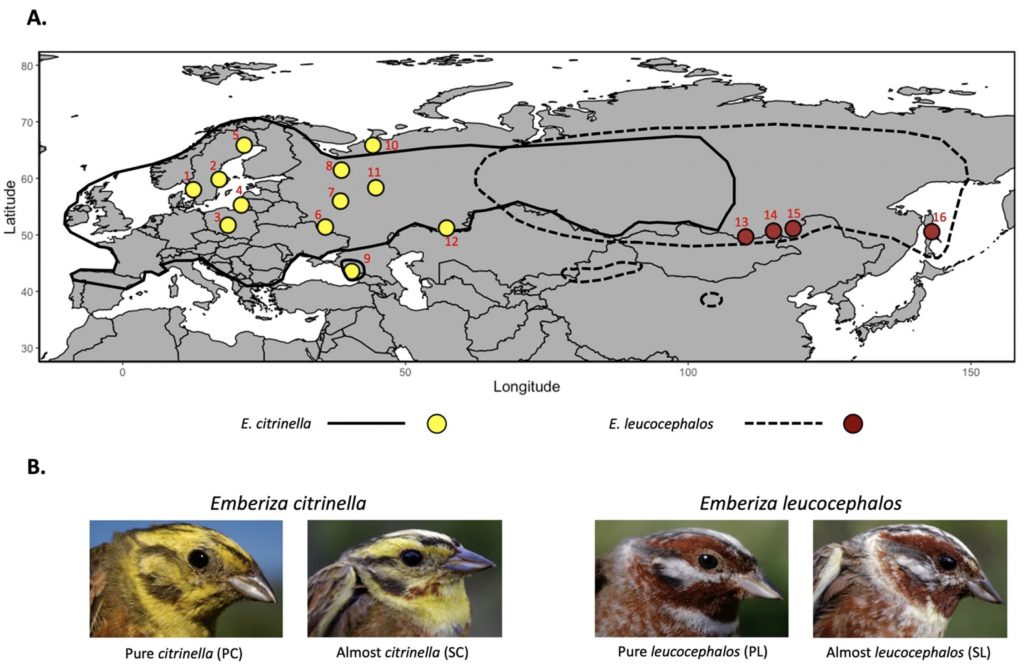
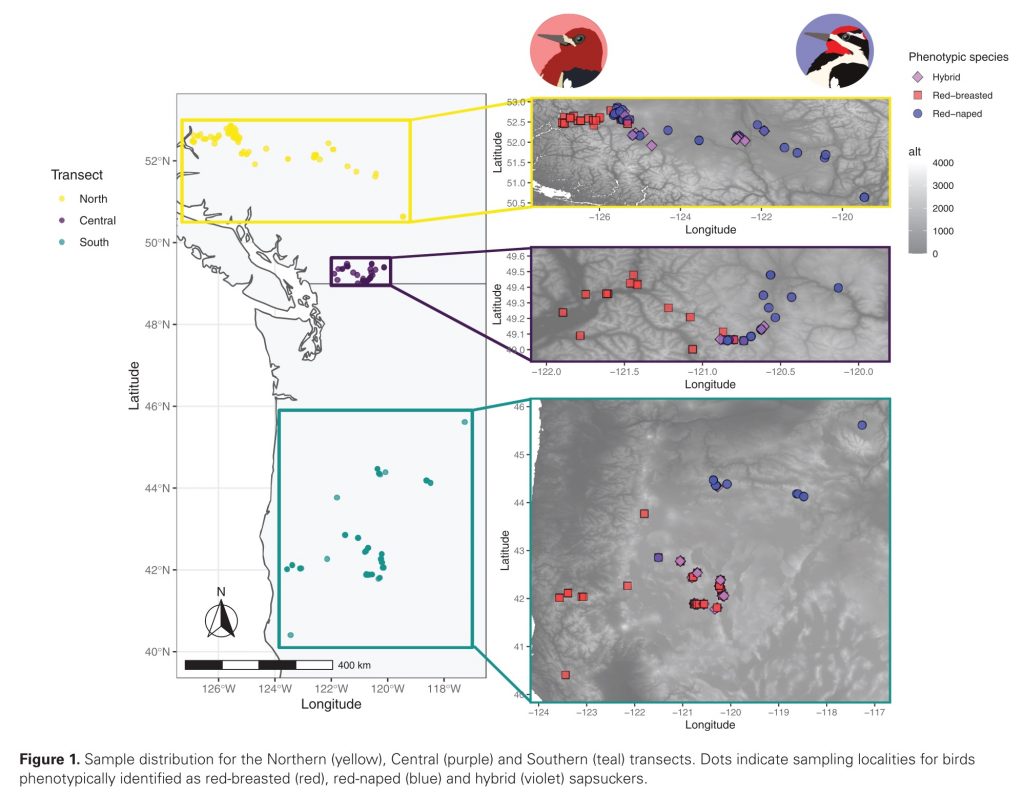
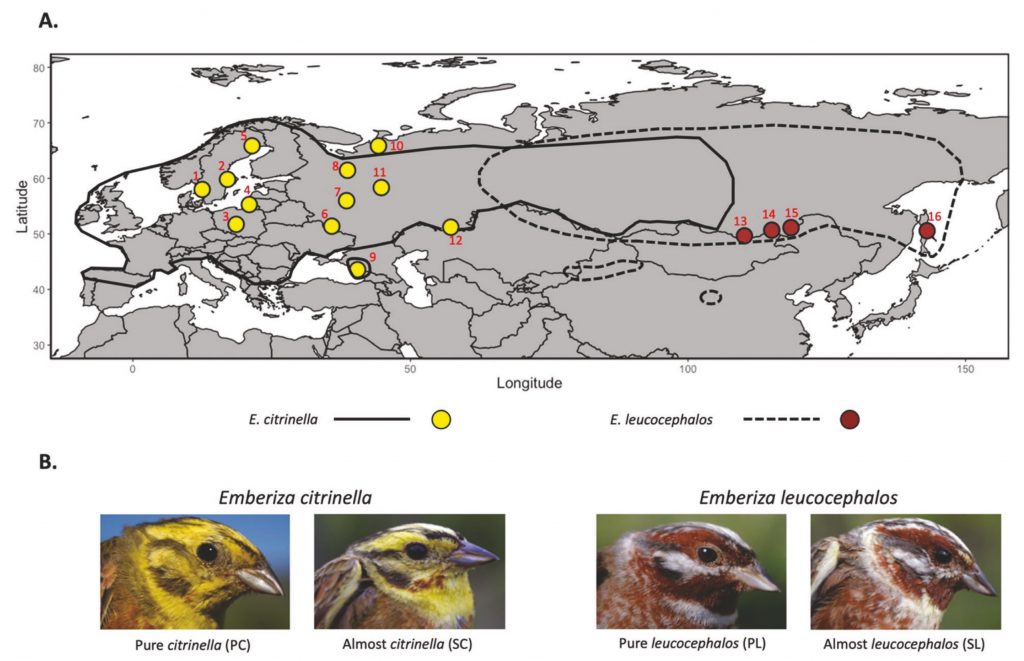
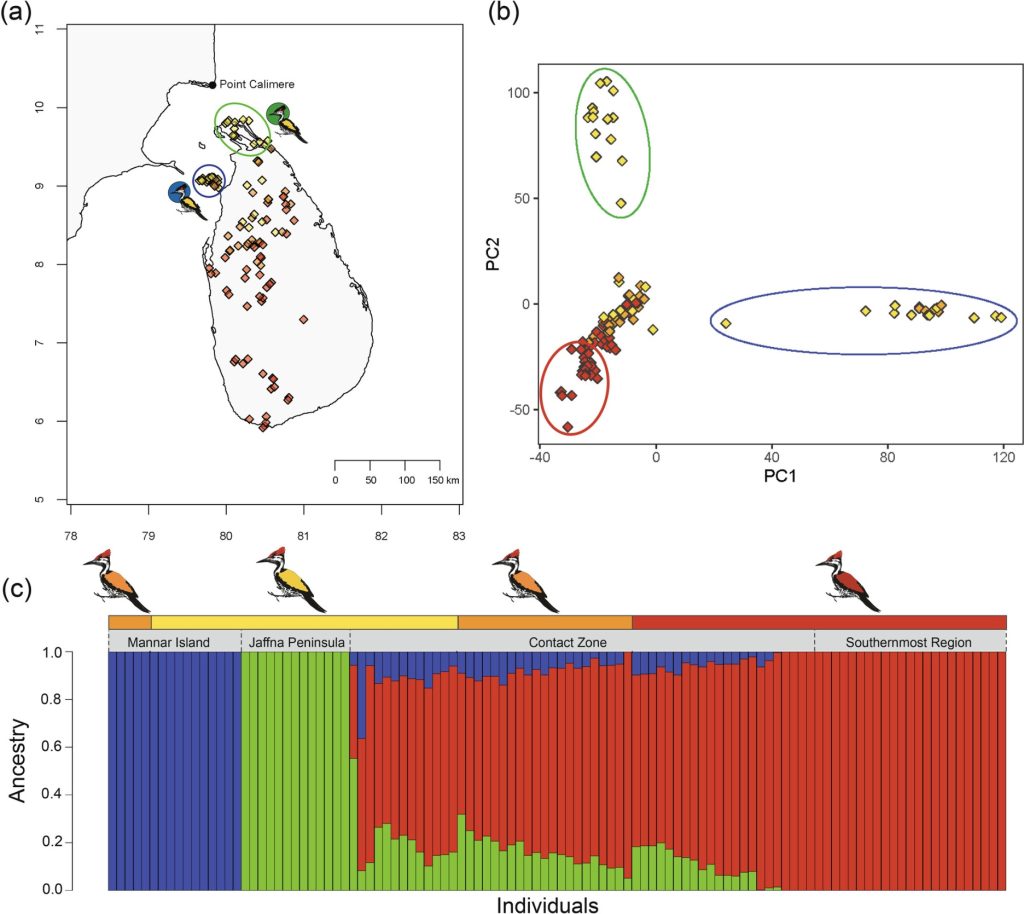
Island ecosystems have emerged as vital model systems for evolutionary and speciation studies due to their unique environmental conditions and biodiversity. This study investigates the population divergence, hybridization dynamics, and evolutionary history of hybridizing golden-backed and red-backed Dinopium flameback woodpeckers on the island of Sri Lanka, providing insights into speciation processes within an island biogeographic context. Utilizing genomic analysis based on next-generation sequencing, we revealed that the Dinopium hybrid zone on this island is a complex three-way hybrid zone involving three genetically distinct populations: two cryptic populations of golden-backed D. benghalense in the north and one island-endemic red-backed population of D. psarodes in the south of Sri Lanka. Our findings indicate asymmetric introgressive hybridization, where alleles from the southern D. psarodes introgress into the northern D. benghalense genome while phenotype remains adapted to their respective northern arid and southern wet habitats. The discovery of two genetically distinct but phenotypically similar D. benghalense populations in northern Sri Lanka highlights the process of cryptic population differentiation within island ecosystems. These populations trace their ancestry back to a common ancestor, similar to the Indian form D. b. tehminae, which colonized Sri Lanka from mainland India during the late Pleistocene. Subsequent divergence within the island, driven by selection, isolation by distance, and genetic drift, led to the current three populations. Our findings provide evidence of cryptic diversification and within-island population divergence, highlighting the complexity of hybridization and speciation processes. These findings further emphasize the intricate nature of evolutionary dynamics in island ecosystems.