Lecture 8 & 9
Transcription Factor Dimerization and its consequences:
Jun and Fos
In the next series of lectures we will discuss some of the solutions to the problems posed in Lecture 7. Namely, we will discuss how transcription factor dimerization plays a key role in gene regulation. Many eukaryotic transcription factors form dimeric (2:1) protein-DNA complexes. Dimers forming between identical proteins are known as homodimers, whereas dimers forming between different proteins are known as heterodimers. Dimerization can act to increase DNA binding affinity (ie. cooperativity), to create novel binding specificity, allows a small protein to recognize a larger DNA sequence, can explain the dual function of some proteins involved in activation and repression, and can explain how different members of gene families show different DNA-binding specificity.
Combinatorial Control
Although embryonic development is a multi-step process characterized by the sequential activation and repression of many genes, only a relatively small number of transcription factors are responsible for regulating the expression of developmental genes. This diversity in gene regulation by a limited number of transcription factors is achieved through a complex network of interactions between these proteins and specific DNA sequences found in enhancers and promoters of developmental genes. The sequence of these DNA elements is critical in determining the composition and architecture of the transcriptional complexes that ultimately control gene expression in a developmental-specific manner. More specifically, the orientation, spacing and slight alteration of these sequences dictate the identity and mode (monomer, heterodimer, or homodimer) of transcription factor binding which ultimately results in diverse effects on transcription. This type of transcriptional regulation is known as combinatorial control and is a very important concept in this course.

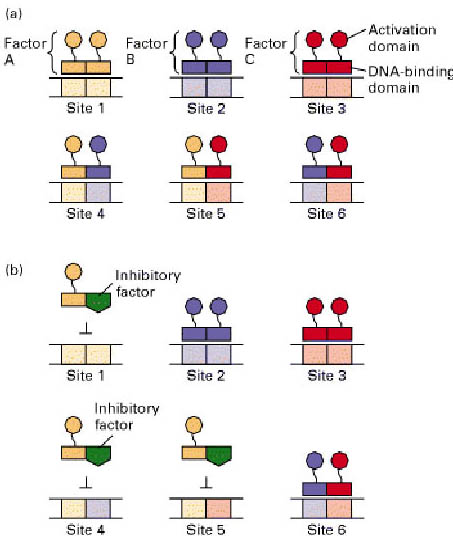
Dimerization of transcription factors allows combinatorial control. (a) In this hypothetical example, transcription factors A, B and C can each form homodimers and heterodimers. This permits the three factors to bind to six different DNA sequences (1-6) and creating six combinations of activation domains. Note that each binding site is divided into two half-sites, and that a single heterodimeric factor contains the activation domains of each of its constituent monomers. (b) When an inhibitory factor (green) is expressed that interacts only with factor A, binding to sites 1, 4 and 5 is inhibited, but binding to sites 2, 3 and 6 is unaffected.
Combinatorial control such as this may provide for tissue-specific gene expression in the absence of tissue-specific transcription factors. Only the combination of proteins needs to be tissue-specific. For example, factor A may be expressed in thymus, brain and liver; factor B may be expressed in thymus, pancreas and heart. The factors are only present together in the thymus and regulate thymus-specific gene expression. In this way, combinatorial control provides a way for a limited number of transcription factors to regulate a large number of genes.
Evidence for dimerization
The first indication that transcription factors dimerize came from DNA footprinting data that showed most transcription factor binding sites are palindromic (inverted repeats). This suggested that mirror-image pairs of proteins were contacting similar DNA surfaces. In most transcription factors the dimerization domain is separable from the DNA-binding domain. Studies in which the putative dimerization domain was mutated demonstrated that the domain is necessary for protein association, and that failure to dimerize also resulted in failure to bind DNA in EMSA or DNA footprinting assays.
Today we will be discussing the following paper:
DNA Binding Activities of Three Murine Jun Proteins: Stimulation by Fos. Y Nakabeppu, K. Ryder and D. Nathans. Cell, Vol. 55, 907-915, December, 1988.
Introduction
Reviewed in 1) Oncogene 2001 Apr 30;20(19):2438-52.
2) Oncogene 2001 Apr 30;20(19):2453-64.
Members of the Fos and Jun protein families are involved in the regulation of a variety of cellular process including cell proliferation, differentiation, apoptosis and oncogenesis. Members of this family include Fos, Fra-1, Fra-2, FosB, Jun, JunB and JunD, are HLH (bZip) proteins and are expressed in numerous cell types. Fos and Jun family proteins function as dimeric transcription factors that bind to AP-1 regulatory regions of many genes. The AP-1 site is an asymmetric heptanucleotide recognition sequence, TGA(C/G)TCA, that is located in a wide range of enhancers and promoters. Dimers formed by Jun and Fos bind with the highest affinity to this consensus AP-1 site and with less affinity to variants thereof. It has been proposed that the variation in the AP-1 sequence contributes to the differential functions of different Jun-Fos family dimers at various regulatory elements.
This paper by Nathans group is a milestone paper in Jun/Fos biology. It examines the Jun family to see if all Jun proteins bound DNA and to see if the Jun family can form homo- and heterodimers. The major finding is that Jun proteins are able to form both homo- and heterodimers. In addition, the Jun proteins are similar in their interaction with Fos proteins that leads to enhanced binding to an AP-1 site in DNA.
Question 1. Do all three Jun family proteins recognize the AP1 site? What experiments would you perform to figure this out? How do you interpret their data?

Question 2. Do all Jun proteins form homodimers? How would you figure this out?


Question 3. Do the Jun proteins form heterodimers? How would you figure this out?

Class Discussion:
How would you show the presence of Fos in the Jun-DNA complex?
Does Fos have an effect on the affinity of Jun-B binding to the AP-1 site?