Lecture 4
Terms and Molecular Techniques
Measuring Gene Expression
In this section I will provide you with information about molecular biology terms and molecular techniques which you should be familiar with. We will see most of the important techniques as we discuss primary research papers. You should be aware that one of the things you need to know is when to apply techniques such that you can design experiments to answer specific biological questions, and know how they work so that you can interpret the data.
Glossary
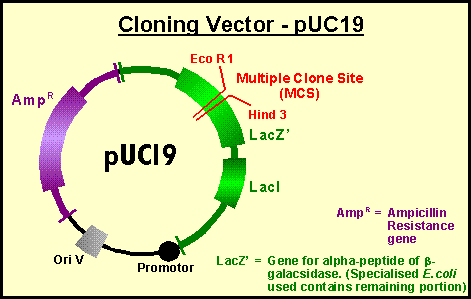
Vector: A self-replicating DNA that carries a selectable marker, a cloning site for insertion of DNA, and may carry other useful fragments of DNA such as promoters, termination sites or reporter genes. Vectors are usually derived from plasmids, phage, or viruses.

Insert: A piece of foreign DNA cloned into a vector.
Clone: 1) noun - vector plus insert. 2) noun - A propagating population of organisms, either single cell or multicellular, derived from a single progenitor cell. Such organisms should be genetically identical, though mutation events may abrogate this. 3) verb - To purify a single piece of DNA and ligate it into a vector.
Subclone: Procedure used to ligate DNA segments in a cell-free system.
Shuttle vector: A vector that can grow in two organisms, typically bacteria and eukaryotic cells. Requires two origins of replication, and a selectable marker for each organism. Essential for carrying out cloning steps in bacteria, and then using the clone to test in appropriate cells (ie. yeast, insect etc.)
Expression Vector: A vector that allows transcription of the insert, under the control of its own or a heterologous promoter. The transcript is usually translated, giving you to a way to make a protein by having a clone for the gene or a cDNA.
Reporter gene: A gene cloned downstream of a heterologous promoter or enhancer whose product is easily measured or identified, and is not normally made in the host cell. This provides a convenient way of quantifying the strength of a promoter or an enhancer (or the effects of mutations made in these regions). Some typical reporters are CAT (chloramphenicol acetyl transferase) and luciferase which are enzymes with quick and easy assays useful in cells. b-galactosidase (LacZ) is commonly used in embryonic assays; it stains a colourless day (X-gal) bright blue. GFP (green fluorescent protein) emits green flourescence when excited with the appropriate wavelength of light thus useful for dectecting time and place of gene expression in a living organism.
Transfection: A method for introducing a clone into tissue culture cells. This may be accomplished by calcium phosphate precipitation, the use of liposomes, and electroporation to name a few. There are two types of transfections: transient transfections in which foreign DNA is introduced and is expressed, but does not integrate into host chromosome and is thus unstable, and stable transfections in which foreign DNA is introduced and integrated into the host chromosome, is stably inherited, and a new cell line is formed and isolated which can be reproducibly analyzed. Transient is faster thus one can test many different constructs, however, it is not likely to include chromatin effects and thus is artificial in terms of chromatin or chromosome context of transcription.
Transformation (Transgenic): Incorporation of foreign DNA into the chromosomes of the germline cells. In Drosophila, we use transposons (P-element transformation) to integrate cloned genes into chromosome of germ line cells by microinjection. This procedure produces flies with the foreign gene in all cells of body and future generations. Integration is random although targeted transposition is becoming more popular.

Transformations in mice utilize homologous recombination. Microinject DNA into male pronucleus of fertilized egg. Homologous recombination between the vector and genomic sequences can replace or more often knock out or disrupt a gene.

Transposon Tagging: Based on making mutations as a consequence of the random insertion of transposable elements within the genome, the transposon can be used to clone out the flanking DNA. This is similar to what we discussed in class known as enhancer traps.
Primers: Short fragments of DNA used to hybridize to DNA or RNA so that a cDNA can be synthesized, either by reverse transcriptase or by PCR.
Reverse Transcriptase: An enzyme that will make cDNA from RNA and a primer.
PCR: Polymerase Chain Reaction. Uses primers, template DNA and a thermostable DNA polymerase to amplify a region of DNA.
Library: A collection of clones containing DNAs from a completely representative collection of clones.
Genomic Library: Collection of phage, cosmids, P1 phagemids or yeast artificial chromosomes containing all the DNA in the genome.
cDNA Library: Collection of clones containing cDNA copies of every mRNA in a given cell, tissue or developmental stage of an organism.
Expression library: A kind of cDNA library in which the cDNAs have been directionally cloned so that each cDNA will be transcribed under the control of an inducible promoter (recall expression vectors).
Antibodies: Antibodies against a given protein are made by injecting a higher vertebrate with purified protein over the course of several months. The animal's immune system will respond by making antibodies directed against that protein. The IgC can be purified from the serum and then the specific antibody can be biochemically purified using the antigen. This is a primary antibody. Usually a secondary antibody is used to detect the primary antibody. So if the primary was generated in rabbits, a different animal's antibodies directed against rabbit IgC will be used as a secondary. For detection, the secondary antibody is conjugated to an enzyme that catalyzes a reaction that can somehow be visualized.
METHODS
Electrophoresis: Electrophoresis is the migration of charged molecules in gels of acrylamide or agarose in response to an electric field. Their rate of migration depends on the strength of the field; on the net charge, size and shape of the molecules and also on the ionic strength, viscosity and temperature of the medium in which the molecules are moving. As an analytical tool, electrophoresis is simple, rapid and highly sensitive. It is used analytically to study the properties of a single charged species, as a separation technique or to estimate the size of a purified molecule. Because dilute agarose gels are generally more rigid and easy to handle than polyacrylamide of the same concentration, agarose is used to separate larger macromolecules such as nucleic acids, large proteins and protein complexes. Polyacrylamide, which is easy to handle and to make at higher concentrations, is used to separate most proteins and small oligonucleotides that require a small gel pore size for retardation.
The electrophoretic mobility of proteins in polyacrylamide gels depends on both the size as well as the charge-to-mass ratio. Therefore, to separate proteins according to size it is necessary to give them the same charge-to-mass ratio. This is accomplished by denaturing the protein in SDS (sodium dodecyl sulfate) hence the term SDS-PAGE. Such gels are stained with protein specific stains (eg. Coomassie Brilliant Blue) to show the position and levels of each separated component (the amount of stain is proportional to the level of expression of the gene specifying that polypeptide).
Measuring Gene Expression
Blotting Methods: To detect a given molecule in an electrophoretically separated mixture, you need a way of detecting the molecule in question. To do so, the molecule(s) must be transferred out of the gel and onto a filter. The transfer of molecules out of the gel and onto the filter conserves the spatial organization of the molecules and can be done by capillary transfer or electrophoretically. The filter is then blocked to prevent non-specific binding, and then challenged with a labelled DNA, RNA to detect DNA or RNA, or with an antibody to detect a specific protein.
|
Name |
Molecule electrophoresed on gel |
Probe |
Application |
|
DNA |
DNA/RNA |
Determine size, presence or absence |
|
|
RNA |
DNA/RNA |
Determine size, distribution, abundance |
|
|
Protein |
Antibody |
Determine size, distribution, abundance |
|
|
South Western |
Protein |
DNA |
Determine size of DNA-binding protein |
|
Far Western |
Protein |
Labelled protein |
Determine size of protein that binds labelled protein |
Determining where and when genes are expressed:
The basic idea is to look for transcripts. This can be accomplished by developmental Northern blots in which RNA from every tissue and developmental stage of interest is purified. This can be done in situ on whole mount embryos/adults or on tissue sections (in situ hybridization). Alternatively, specific proteins extracted from various tissues at developmental stages can be detected on western blots, or using antibodies on whole mounts or sections. The problem with protein detection is that you can't tell newly synthesized protein from lingering protein made earlier. Each of these methods will be discussed in turn below.
Assays for mRNA product:
1. In vitro translation
The in vitro synthesis of proteins in cell-free extracts has a variety of applications, including the characterization of unknown gene products, the generation of labelled protein free of other contaminants, localization of mutations through synthesis of truncated gene products, protein folding studies, and incorporation of mutations for functional studies.
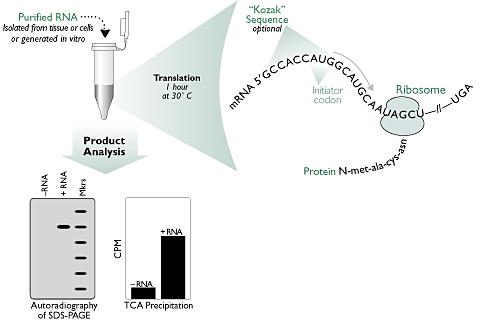
http://www.ambion.com/basics/index.html
2. Northern Blots (Dot blotting)
RNA dot/slot blots: These methods allow the rapid analysis of numerous small samples for the sequence of interest and is less time consuming than the gel electrophoresis methods. 'Dots' or 'slots' of RNA are made onto a filter using a manifold and the filter is then hybridized with a labeled probe.
Developmental northern blots:

Horowitz and CA Berg (1996). Development 122: 1859-1871.
3. Nuclear Run-off (or run-on): This is a system used to study primary rate of transcription. Active nuclei isolated from tissues are supplied with radioactive ribonucleotides and allowed to complete transcripts already started. Specific transcripts (from expressed genes) can be detected and quantified by hybridizing to cloned DNA immobilized on a membrane (dot blot).
4. In situ hybridization: Labeled DNA or RNA is used to detect the complementary mRNA in whole mount embryos or tissue sections - labels the cells in which the gene is being transcribed and therefore where the product is synthesized.

5. Quantitative Reverse Transcription PCR (RT-PCR): RT-PCR is becoming an increasingly popular method for the quantitative analysis of gene expression. This method is used where quantitation is impossible by other means, such as the study of low abundance RNAs or in studies where the amount of starting material is low. Use of an internal standard amplified by the same set of primers as the template RNA allows for great accuracy.

Assays for Protein Quantitation and Localization:
1. Enzyme Assays: Enzyme activity measured in protein extracts from tissues; colorimetric reactions using substrates that produce a detectable product (ie. colour reaction) or radioactive substrates. Measurement of any biochemical reaction in which product appearance or substrate disappearance can be measured. These assays are often combined with gel electrophoresis to separate protein components.
2. Immunoassays: ELISA (enzyme-linked immunosorbent assay) is performed on protein extracts using antibodies conjugated to enzymes which can be assayed with a colorimetric or fluorimetric substrate. In most cases, colour is proportional to amount of protein product. Often used in combination with electrophoresis; immunoelectrophoresis to quantify specific proteins separated by electrophoresis.
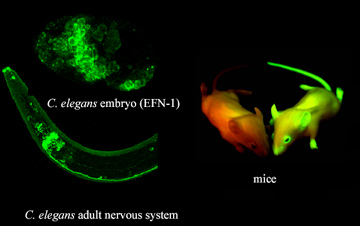
3. Immunolocalization: Enzyme-conjugated or labelled antibodies used to detect specific proteins in tissue sections or whole mounts to identify when and where proteins of interest are expressed. Intensity of signal is proportional to amount of protein product. Similarly, fusion of regulatory sequences to the green fluorescent (GFP) reporter gene results in the deposition of GFP in those cells where the gene is transcribed making it an ideal in vivo marker for gene expression.

4. Protein gel electrophoresis: See section on gel electrophoresis.
5. Western blotting: Proteins separated by electrophoresis, blotted onto membrane and components detected using specific antibodies. Protein may be quantified by comparison to internal standards.
