![]()
![]()
Evolution
A cumulative change in the characteristics of organisms or populations from generation to generation.(Academic Press Dictionary)
Natural selection
The process whereby some individuals contribute more offspring to the next generation as a consequence of their carrying a trait or traits favorable to survival or reproduction.
Artificial selection
The process of selection whereby traits in an organism are deemed favorable and are selectively bred by humans. (Humans specify who survives and reproduces.)
Homology
A similarity between species that is not functionally necessary but that results from inheritance from a common ancestor.
Vestige
A bodily part or organ that is degenerate or imperfectly developed in comparison to one more fully developed in closely related forms. (Webster's Dictionary)
![]()
Numerous lines of evidence exist for evolution. These may be categorized as follows (following Ridley):
1. Direct observation of change in natural populations
2. Direct observation of change under artificial selection
3. Homologous traits
4. Homologies may be hierarchically classified (nested)
5. Evidence for evolutionary change in fossil record
![]()
![]()
HIV is the retrovirus that causes acquired immune deficiency syndrome (AIDS) in humans.
As a retrovirus, the virus particle (called a virion) contains RNA not DNA. When a retrovirus infects a host cell (1), the RNA becomes translated into DNA (2-3) by a protein called "reverse transcriptase" that is encoded in the viral RNA.
This DNA then enters the nucleus and integrates into the host cell's nuclear genome at a site that is actively transcribed (4). The DNA of the virus is subsequently transcribed and translated by the host cell (5), producing both the RNA and proteins needed to produce new virions (6).
Finally, new virions burst out of the host cell (7).

HIV contains only nine genes. We'll discuss the evolution of two of these genes: env (the gene that produces the outer surface glycoproteins of HIV) and pol (the gene that produces reverse transcriptase).

HIV has a high mutation rate and evolves very rapidly, thwarting the defenses of the immune system and the efficacy of drug therapy.
![]()
Ganeshan et al. (1997) sequenced regions of the env gene from six children who were infected with HIV during pregnancy. Sequences were determined at four different time points from ~12 viral clones each time.
Initially, all viruses within a child were similar. Over time, however, the viral sequences changed.
Ganeshan et al. illustrated these changes using a phylogenetic tree, which places sequences that are similar to one another close together. [Branch distances between two sequences on the tree is related to the number of sequence differences between them.]

Observations:
This phylogeny demonstrates that each child was infected by a slightly different HIV virus and that the viral population then changed and diverged within each child (=evolution).
But why?
Interestingly, children D-F were slow to progress to disease, suggesting that they had mounted an adequate immune response to the virus.
In these children, the virus appears to have evolved at a higher rate (witness the longer branches).
Two possible explanations: A higher mutation rate. Selection.
How could we tease these apart?
Genetics review:
Mutations within a gene can be classified as:
If originally a DNA codon is CTC (coding for glutamic acid), a mutation in the third position to CTT would still code for glutamic acid (SYNONYMOUS), but a mutation to CTA would code for aspartic acid (NON-SYNONYMOUS).
If all mutations are neutral (not selected), then synonymous and non-synonymous mutations should occur at the rate expected given the genetic code.
If changing the amino acid of a protein is detrimental to the functioning of the protein, however, non-synonymous mutations would be eliminated by selection and would be less likely to remain within the population.
![]() Non-synonymous mutations less frequently observed.
Non-synonymous mutations less frequently observed.
If changing the amino acid of a protein is beneficial, however, non-synonymous mutations would be promoted by selection and would be more likely to remain within the population.
![]() Non-synonymous mutations more frequently observed.
Non-synonymous mutations more frequently observed.
Ganeshan et al. found that the rate of non-synonymous mutations in children D-F was significantly higher than the rate of synonymous mutations.
This suggests that, in these children, natural selection actively favored mutations in the env gene that changed the amino acid sequence of the viral surface proteins.
This is consistent with the hypothesis that children who are slow to progress to AIDS have an active immune system.
Active immune systems strongly select for HIV viruses that evolve different surface proteins, thus evading the immune system.
In the children who progressed to AIDS rapidly, Ganeshan et al. observed the opposite pattern. This suggests that the virus was already capable of evading the immune system and was not under selection to change.
![]()
Azidothymidine (AZT) is a drug designed to combat HIV by mimicking the nucleotide thymidine (T).
Reverse transcriptase uses AZT instead of T when translating RNA into DNA, which then blocks the growing DNA chain.
Initially, AZT was quite effective at halting the deterioration of the immune system in people with AIDS.
Within a few years, however, AZT stopped working in many of these patients.
Researchers found that the gene that encodes reverse transcriptase (pol) had changed over time (= evolved) within these patients.
The changes conferring resistance to AZT altered amino acids in the active site of the reverse transcriptase enzyme.
Often, the same changes occurred in different patients (= convergent evolution).
HIV replicates about 300 times per year, producing a large population of virus particles, many of which carry new mutations.
Viruses that contain mutations that decrease the affinity of reverse transcriptase for AZT are much more likely to replicate.
![]() In the presence of AZT, natural selection favors those variants of HIV that are least likely to use AZT when translating RNA into DNA.
In the presence of AZT, natural selection favors those variants of HIV that are least likely to use AZT when translating RNA into DNA.
We can't stop HIV from evolving, but what might slow down the evolution of HIV?
SOURCES:
![]()
![]()
|
|
13 species of finches live on the Galapagos Islands. These species do not differ much in body shape or coloration, and are difficult to distinguish by eye. However, they differ dramatically in beak size and shape. We'll focus on the medium ground finch, Geospiza fortis. These finches feed on seeds, and the size and shape of their beaks determines which seeds they can eat. Birds with deep beaks are able to crack large, hard-shelled seeds, while birds with shallow beaks are better at feeding on small, soft seeds. So, the distribution of seeds is an important component of the environment.
|
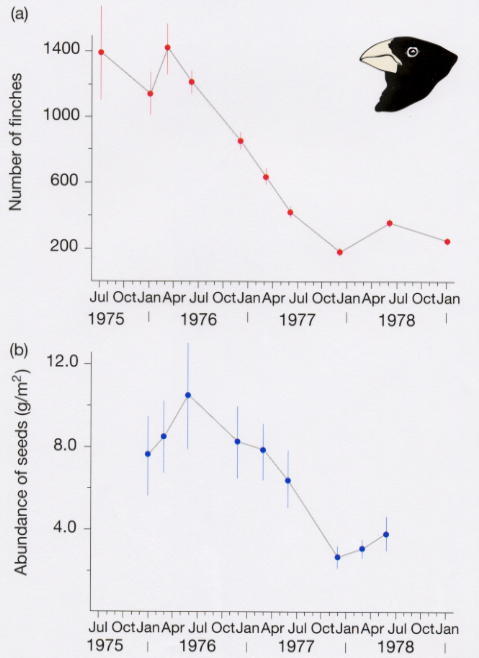
During severe weather brought on by an El Nino in 1977, food became quite scarce and many finches died. The population on one island went from 751 birds to 90 birds. This was an episode of extremely strong selection.
Very few small seeds were available. Many remaining seeds were very hard-shelled and could only be cracked by birds with large, deep beaks. So, the depth of a bird's beak became a trait very important to its survival. Birds with deeper beaks were more likely to survive the El Nino.

Before the 1977 El Nino, average beak depth was about 9.4 mm. After the El Nino, the average beak depth of surviving birds was about 10.2 mm. This change in beak depth can be directly attributed to the abundance of seeds and the ability of birds to eat the seeds that were available.
|
|
![]()
![]()
Cody and Overton (1996) studied changes in seed size and shape that occurred in weedy, wind-dispersed plants located on the islands off the westcoast of Vancouver Island.

For eight summers between 1981 and 1991, Cody and Overton censused the plant populations of 200 islands and a region of the mainland.
Extinction and recolonization events occurred frequently on the islands.
For a wind-dispersed plant, how might seeds that successfully colonize an island differ from the majority of seeds on the mainland?
After a plant has colonized an island, how might seed morphology evolve over time?
Cody and Overton studied two species that were common and whose seed dispersal was directly related to seed morphology: Hypochaeris radicata and Lactuca muralis.
|
Both are asteraceae that produce achenes consisting of a feathery pappus and a seed. |
|
Dispersal ability of the achenes in these two species is related to the volume of the pappus (VP) divided by the volume of the achene (VA).
|
Old island populations of Hypochaeris radicata have a significantly (p<0.01) lower dispersal ability than mainland populations. |
|
For Lactuca muralis, enough extinction and recolonization events have been observed to break down the changes in seed shape by the number of years that the plant has been on an island.

As predicted, plants that have recently arrived on an island produce seeds with greater dispersal ability than the mainland, but dispersal ability significantly decreases (p<0.01) as the age of the population increases.
The seed morphology of the island plant populations studied by Cody and Overton have evolved in a manner consistent with strong natural selection for dispersal ability among colonists and against dispersal ability among residents of these islands.
SOURCES: