Lecture 16
Anterior-Posterior axis formation in Drosophila
The process of segmentation along the anterior-posterior axis of Drosophila is controlled by the hierarchical interaction between five sets of genes. These are the maternal-effect, gap, pair-rule, segment-polarity and the homeotic genes. We have briefly discussed these group of genes in class but it is my intention, in the next series of lectures, to teach you the molecular mechanisms involved in the generation of the anterior-posterior axis in the Drosophila embryo.
Morphogen gradients:

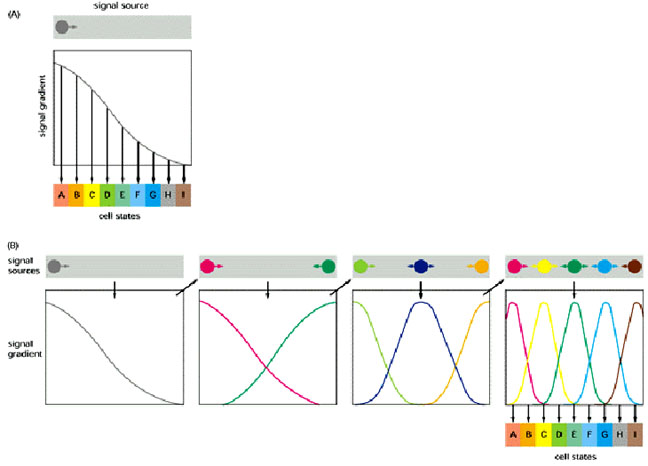
Two strategies for using morphogen gradients to specify a fine-grained pattern of cells in different states. (A) If there is only one morphogen gradient, cells select their states by responding to small changes of signal concentration. (B) The initial morphogen gradient(s) control the establishment of a small number of more local signals, which control the establishment of other, more refined, local signals. Because there are multiple local signals, the cells do not have to respond very precisely to any single signal in order to creat the correct spatial array of cell states. Case B corresponds more closely to what happens in the Drosophila embryo and can be represented by the following figure which illustrates combinatorial control:
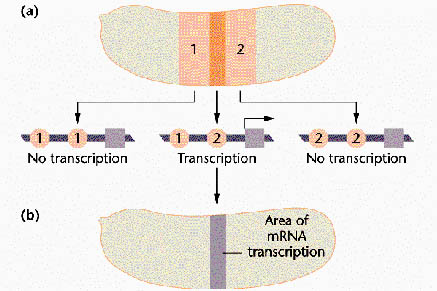
The Gap genes:
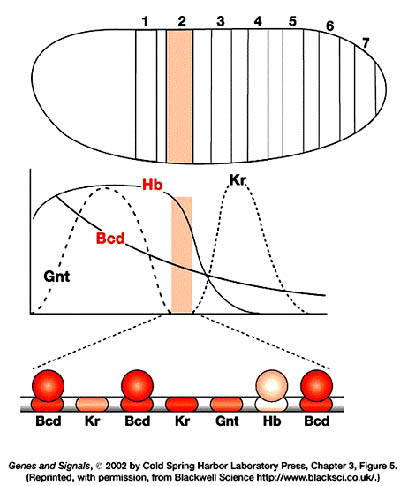
Gap genes participate in early patterning along the anterior-posterior axis of Drosophila embryos. These zygotically transcribed genes, including hunchback, Krüppel, knirps and giant, are expressed in specific spatial domains within 2 hours AEL, just before the cellular blastoderm stage. Genetic and molecular experiments suggest that the boundaries of expression of Krüppel, Knirps and Giant reflect a balance of transcriptional activation and repression involving maternally derived Bicoid and Nanos as well as Hunchback, the first gap gene to be transcribed. In fact, Bicoid strongly activates hunchback transcription and then binds cooperatively with Hunchback protein to several target gap genes, such as orthodenticle, giant and hunchback itself; as a result, the target genes are activated in specific domains. High concentrations of Hunchback protein repress transcription of Krüppel, but below a critical threshold concentration, Hunchback acts as a transcriptional activator of Krüppel. This threshold establishes the anterior boundary of the Krüppel expression domain. More posteriorly, the Hunchback concentration falls below the threshold at which it activates transcription of Krüppel, thereby setting the posterior Krüppel boundary. Expression in their anterior domains is activated by Bicoid, while expression in their posterior domains is activated by the combined action of Bicoid and Nanos. The anterior boundaries of the Knirps and Giant expression domains are determined by Hunchback-mediated transcriptional repression, and their posterior boundaries are determined by the product of an additional gap gene, tailless.
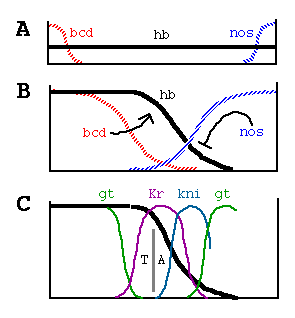
From Struhl et al. Control of Drosophila Body Pattern by the hunchback Morphogen Gradient. Cell, Vol. 69, 237-249, April 17, 1992.
Generation and Function of the
Hunchback Morphogen Gradient
(A) bicoid (bcd) and nanos (nos) maternal
transcripts are tightly localized at the anterior and posterior poles of the
unfertilized egg, in contrast to hunchback (hb) transcripts,
which are ubiquitously distributed.
(B) Shortly after fertilization, bcd transcripts are translated, and the
resulting protein diffuses posteriorly, generating a gradient; similarly, nos
transcripts are thought to generate an opposing gradient of nos activity,
presumably its encoded protein. bcd activates zygotic hb transcription
anteriorly, whereas nos translationally represses hb transcripts
posteriorly. As a consequence, Hb protein accumulates differentially, declining
in a graded fashion from high, uniform levels at the anterior end to
undetectable levels at the posterior end. (C) The Hb protein gradient then
provides a series of concentration thresholds that independently dictate where
the anterior Kr, kni, and gt boundaries, as well as the posterior Kr boundary,
are positioned in the posterior half of the body. The posterior kni boundary is
governed in part by gt (and hence indirectly by hb ) and by the terminal system,
which also controls the posterior gt boundary. The domain of anterior gt
expression (activated under bcd control) also depends on the concentration of hb
protein. Finally, the hb gradient dictates the boundary between thorasic (T) and
abdominal (A) differentiation, possibly by directly repressing bithorax complex
gene activity.
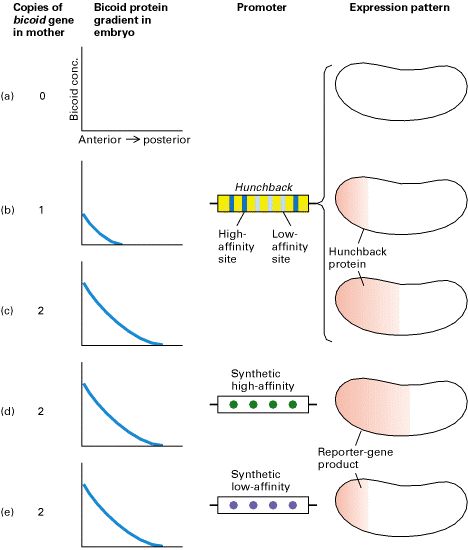
Experimental demonstration that transcription of target genes, such as hunchback, directly regulated by Bicoid protein reflects the concentration of Bicoid and its affinity for protein-binding sites in the promoter of the target gene. (a-c) Increasing the number of bicoid genes in mother flies changed the Bicoid gradient in the early embryo, leading to a corresponding change in the gradient of Hunchback protein (red) expressed from the zygotic hunchback gene. (d and e) The hunchback promoter has been shown to contain three high-affinity and three low-affinity Bicoid-binding sites. Transgenic flies carrying a reporter gene linked to a synthetic promoter containing either four high-affinity sites (d) or four low-affinity sites (e) were prepared. Expression of the reporter-gene product was dependent on the affinity of the Bicoid-binding sites in the promoter. [Adapted from D. St. Johnston and C. Nüsslein-Volhard, 1992, Cell 68:201.]
The Pair-rule genes:
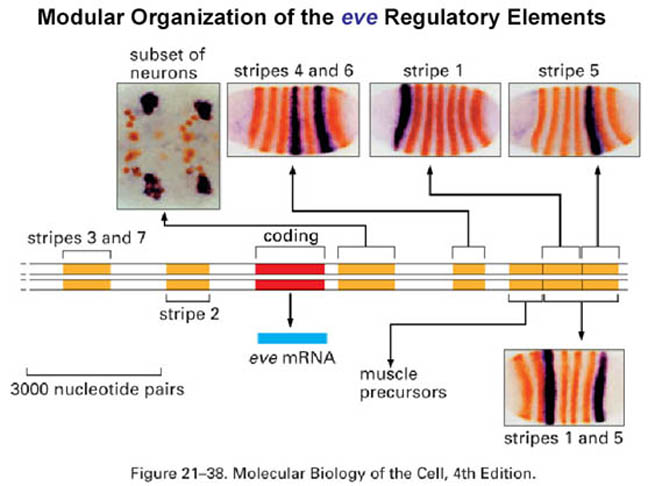
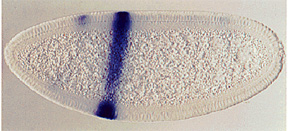
The pair-rule genes, which include fushi tarazu, hairy, and even-skipped, encode transcription factors that are expressed in stripes of cells, corresponding to the parasegments, covering the central part of the embryo. Each pair-rule gene product is expressed in seven parasegments, either the even or odd numbered.

The pair-rule genes can be subdivided based on their mode of regulation. The different stripes of the primary pair-rule genes, which include even-skipped, hairy and runt, are independently regulated through enhancer elements called stripe initiation elements. This means that separate enhancer elements control the expression of each stripe of gene expression. Regulation of gene expression in each stripe is achieved through overlapping subsets of maternal-effect and/or gap gene products that either activate or repress a particular stripe element.
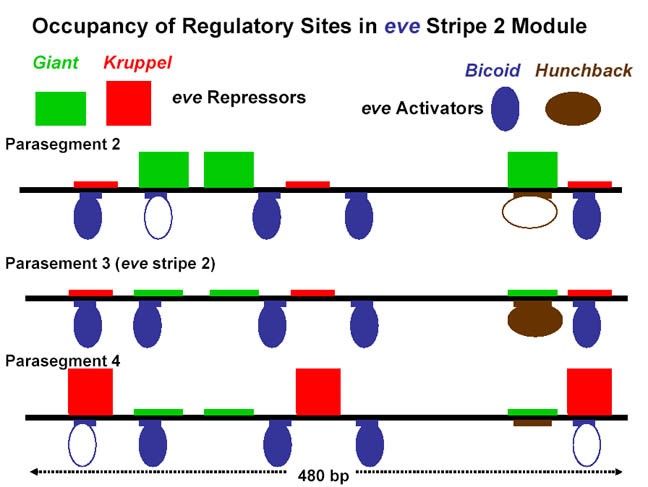
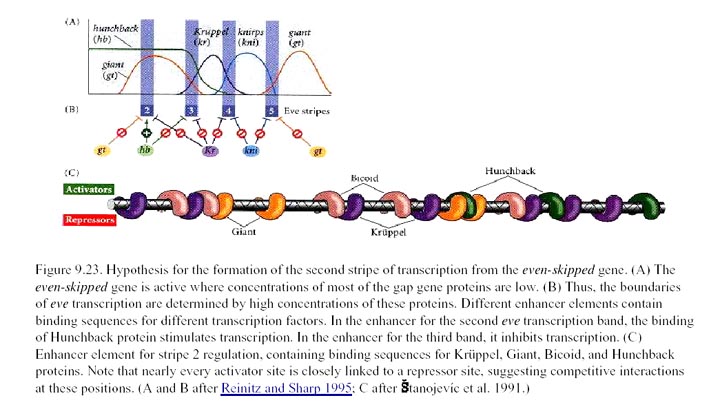
 stripe 2 and of the four proteins that regulate its expression. The
coordinated effect of the two repressors (↓)
and two activators (↑) determine the precise boundaries of the second
anterior Eve stripe. Expression of
other stripes is regulated independently by other
combinations of transcription factors encoded by maternal and gap genes.
Below that is a simplified diagram of the 815-bp regulatory region controlling
transcription of the stripe 2 of eve. This region contains binding
sites for Bicoid and Hunchback
proteins, which activate transcription of eve, and for Giant and Krüppel
proteins, which repress transcription. [We will be discussing the
experiments leading to these findings in the next lecture. See S. Small
et al., 1991, Genes & Devel. 8:827.]
stripe 2 and of the four proteins that regulate its expression. The
coordinated effect of the two repressors (↓)
and two activators (↑) determine the precise boundaries of the second
anterior Eve stripe. Expression of
other stripes is regulated independently by other
combinations of transcription factors encoded by maternal and gap genes.
Below that is a simplified diagram of the 815-bp regulatory region controlling
transcription of the stripe 2 of eve. This region contains binding
sites for Bicoid and Hunchback
proteins, which activate transcription of eve, and for Giant and Krüppel
proteins, which repress transcription. [We will be discussing the
experiments leading to these findings in the next lecture. See S. Small
et al., 1991, Genes & Devel. 8:827.]
Expression of the secondary pair-rule genes is dependent on the prior expression of the primary pair-rule genes. Furthermore, expression of the secondary pair-rule genes is dependent on single regulatory elements. For example, a 669bp regulatory region of ftz, called the zebra element, contains binding sites for a multitude of activating and repressing transcription factors.
The segment-polarity genes:
The pair-rule
genes act combinatorially (several of their proteins are expressed within a
given cell) to regulate the transcription of the segment-polarity genes,
which are expressed in offset patterns of 14 stripes. Thus, the hierarchy of
transcription-factor regulation extends all the way from the
positional-information system to the repeating pattern of segment-polarity gene
expression. The products of the segment-polarity genes then permit the 14
segments to form and define the individual A![]() P
rows of cells within each segment.
P
rows of cells within each segment.